Datamodel¶
Introduction¶
Containers¶
The AMUSE datamodel is based on objects stored in containers. The containers store all the relevant data of the objects. One can work with the data in a container as a whole or with an individual object in a container. When working with an individual object all data will be retrieved from and stored in the container of that object. This is different from normal python objects and lists where the lists store references to the objects and the data is stored on the objects.
Amuse containers:
>>> from amuse.datamodel import Particles
>>> from amuse.units import units
>>> stars = Particles(2)
>>> stars.mass = [2, 3] | units.MSun
>>> sun = stars[0]
>>> sun.mass = 1 | units.MSun
>>> print stars.mass
[1.0, 3.0] MSun
>>> stars.mass = [0.5, 5] | units.MSun
>>> print sun.mass
0.5 MSun
Python lists:
>>> from amuse.datamodel import Particle
>>> sun = Particle(mass = 1 | units.MSun)
>>> star = Particle(mass = 3 | units.MSun)
>>> stars = [sun, star]
>>> print stars.mass # cannot access attributes on the list
Traceback (most recent call last):
AttributeError: 'list' object has no attribute 'mass'
Set or grid¶
AMUSE supports two kinds of containers. One container kind implements a one dimensional set of objects. You can add and remove objects from the set and combine sets in different ways. This container kind is called ‘Particles’ and the individual objects stored are called ‘Particle’. The other container kind implements a multidimensional grid of objects. You cannot change the shape of a grid and objects cannot be added or removed from a grid. This container kind is called ‘Grid’ and individual objects stored in the container are called ‘GridPoint’.
With particles:
>>> from amuse.datamodel import Particles
>>> from amuse.units import units
>>> stars = Particles(2)
>>> stars.mass = [2, 3] | units.MSun
>>> print stars.mass
[2.0, 3.0] MSun
With grids:
>>> from amuse.datamodel import Grid
>>> from amuse.units import units
>>> field = Grid(2,2)
>>> field.density = [[1, 2], [3, 4]] | units.g / units.cm ** 3
>>> point = field[1,0]
>>> point.density = 5 | units.g / units.cm ** 3
>>> print field.density
[[ 1. 2.], [ 5. 4.]] g / (cm**3)
Memory, code or file¶
The containers in AMUSE can save the data for individual objects in different kinds of ‘Stores’. AMUSE currently implements 3 kinds of stores. One kind of store saves all data in memory, this kind is used by default and provides the fastest way to work with the containers. Another kind of store saves and retrieves all data from a community code. It uses MPI messages to communicate this data. This kind is used by default for containers provided by a code and is the primary way by which you can interact with the data inside a running code. The last kind of container stores and retrieves data from an HDF5 file. This kind is only used when saving or loading a container.
Varying attributes¶
In memory containers can support a user defined set of attributes for the contained objects. You can define a new attribute for the container by assigning a value to the attribute. In code containers support a pre-defined, per code set of attributes for the contained objects. You cannot define a new attribute on these containers. Also as a code may not allow some attributes to be set individually, the container also cannot set some attributes individually. For example you often cannot set the X position of a particle, you must set the X, Y, and Z position in one go.
Adding attributes to a set or object:
>>> from amuse.datamodel import Particles
>>> from amuse.units import units
>>> stars = Particles(keys = [1,2])
>>> # you can add an attribute by assigning to a name on a set
>>> stars.mass = [1, 3] | units.MSun
>>> sun = stars[0]
>>> print sun
Particle(1, mass=1.0 MSun)
>>> sun.radius = 1 | units.RSun
>>> print sun
Particle(1, mass=1.0 MSun, radius=1.0 RSun)
Objects not classes¶
Different kinds of particles or gridpoints are not modelled by implementing subclasses. Instead, different kinds of particles are defined ad-hoc, by variable naming (for example planets, stars or cloud_particles) and varying attributes (stars have position and mass, cloud_particles have position and density). This allows you to model your problem with the names and attributes that best fit your model. Unfortunately, this way of working does remove a level of self description in the system. To mitigate this problem the containers defined in community codes and in example scripts all follow the same conventions. We describe these conventions in a later section in this chapter.
Different attributes and names for different kinds:
>>> from amuse.datamodel import Particles
>>> from amuse.units import units
>>> # stars and planets share some attributes (radius)
>>> # but also have attributes that make sense only for
>>> # the specific kind (population for planets,
>>> # luminosity for stars)
>>> stars = Particles(keys = [1,2])
>>> stars.luminosity = [1, 3] | units.LSun
>>> stars.radius = 1 | units.RSun
>>> planets = Particles(keys = [3,4])
>>> planets.radius = 6371 | units.km
>>> planets.population = [7000000000, 0]
Identity¶
Each object in a container has a unique identity, no two objects in a container can have the same identity. In a ‘Particles’ container this identity is provided by a unique 64-bit key. In a ‘Grid’ container this identity is provided by the n-dimensional index in the grid. Objects with the same identity can exists in multiple containers. These objects are considered as the same conceptual object in AMUSE. Different containers will provide different information about that object. For example the same star could live in a container in memory, in a container of a stellar evolution code and in a container of a stellar dynamics code. AMUSE provides several functions to link these objects and to transfer data between them.
Different sets store information on the same object:
>>> from amuse.datamodel import Particles
>>> from amuse.units import units
>>> stars = Particles(keys = [1,2])
>>> stars.luminosity = [1, 3] | units.LSun
>>> bodies = Particles(keys = [1,2])
>>> bodies.mass = [1, 3] | units.MSun
>>> print bodies[0] == stars[0] # the same 'conceptual' object in different sets
True
>>> print bodies[0] == stars[1] # not the same object
False
>>> print bodies[0]
Particle(1, mass=1.0 MSun)
>>> # finding the coresponding particle in the stars
>>> print bodies[0].as_particle_in_set(stars)
Particle(1, luminosity=1.0 LSun)
Ownership¶
Objects in a container are owned by that container, the container controls the data and the life-cycle of each object.
Particle keys¶
All particles have a unique 64-bit key. This key is created using a random number generator. The chances of duplicate keys using 64-bit integers are finite but very low. The chance of a duplicate key can be determined by a generalization of the birthday problem.
Duplicate keys:
>>> # given n random integers drawn from a discrete uniform distribution
>>> # with range [1,highest_integer], what is the probability
>>> # p(n;highest_integer) that at least two numbers are the same?
>>> import math
>>> number_of_bits = 64
>>> highest_integer = 2**number_of_bits
>>> number_of_particles = 1000000.0 # one million
>>> probability = 1.0 - math.exp( (-number_of_particles * (number_of_particles - 1.0))/ (2.0* highest_integer) )
>>> print probability
2.71050268896e-08
>>> # can also set the probablity and determine the set size
>>> probability = 0.00001 # 0.001 percent
>>> number_of_particles = math.sqrt(2 * highest_integer * math.log(1 / (1.0 - probability)))
>>> print number_of_particles
19207725.6894
If you use large sets or want to load a lot of simulations with
different particles into a script the probability of encountering a
duplicate may be too high. You can check for duplicates in a set of
particles by calling has_duplicates on a set. You can also change
the key generator to better match your problem.
Sets of particles¶
The AMUSE datamodel assumes all particles come in sets. The data of a particle is stored in the set.
This module provides access to all set handling in AMUSE. The actual implementation is in the base, storage and particle modules.
- class amuse.datamodel.AbstractParticleSet(original=None)¶
Abstract superclass of all sets of particles. This class defines common code for all particle sets.
Particle sets define dynamic attributes. Attributes can be set and retrieved on the particles using common python syntax. These attributes can only have values with units.
>>> particles = Particles(2) >>> particles.mass = [10.0, 20.0] | units.kg >>> particles.mass quantity<[10.0, 20.0] kg>
>>> particles.mass = 1.0 | units.kg >>> particles.mass quantity<[1.0, 1.0] kg>
Particle sets can be iterated over.
>>> particles = Particles(2) >>> particles.mass = [10.0, 20.0] | units.kg >>> for particle in particles: ... print particle.mass ... 10.0 kg 20.0 kg
Particle sets can be indexed.
>>> particles = Particles(3) >>> particles.x = [10.0, 20.0, 30.0] | units.m >>> particles[1].x quantity<20.0 m>
Particle sets can be sliced.
>>> particles = Particles(3) >>> particles.x = [10.0, 20.0, 30.0] | units.m >>> particles[1:].x quantity<[20.0, 30.0] m>
Particle sets can be copied.
>>> particles = Particles(3) >>> particles.x = [10.0, 20.0, 30.0] | units.m >>> copy = particles.copy() >>> particles.x = 2.0 | units.m >>> particles.x quantity<[2.0, 2.0, 2.0] m> >>> copy.x quantity<[10.0, 20.0, 30.0] m>
Particle sets can be added together. Attribute values are not stored by the resulting subset. The subset provides a view on two or more sets of particles. Changing attributes of the sum of sets will also change the attributes of each original set, contrary to copy().
>>> particles = Particles(4) >>> particles1 = particles[:2] >>> particles1.x = [1.0, 2.0] | units.m >>> particles2 = particles[2:] >>> particles2.x = [3.0, 4.0] | units.m >>> new_set = particles1 + particles2 >>> print len(new_set) 4 >>> print new_set.x [1.0, 2.0, 3.0, 4.0] m
Particle sets can be subtracted from each other. Like with addition, attribute values are not stored by the resulting subset.
>>> particles = Particles(4) >>> particles.x = [1.0, 2.0, 3.0, 4.0] | units.m >>> junk = particles[2:] >>> new_set = particles - junk >>> print len(new_set) 2 >>> print new_set.x [1.0, 2.0] m >>> print particles.x [1.0, 2.0, 3.0, 4.0] m
Particle sets can have instance based or global vector attributes. A particle set stores a list of scalar values for each attribute. Some attributes are more naturally accessed as lists of vector values. Once defined, a particle set can convert the scalar values of 2 or more attributes into one vector attribute.
>>> from amuse.datamodel import particle_attributes >>> particles = Particles(2) >>> particles.x = [1.0 , 2.0] | units.m >>> particles.y = [3.0 , 4.0] | units.m >>> particles.z = [5.0 , 6.0] | units.m >>> particles.add_vector_attribute("p", ["x","y","z"]) >>> particles.p quantity<[[1.0, 3.0, 5.0], [2.0, 4.0, 6.0]] m> >>> particles.p[0] quantity<[1.0, 3.0, 5.0] m> >>> particles.position # "position" is a global vector attribute, coupled to x,y,z quantity<[[1.0, 3.0, 5.0], [2.0, 4.0, 6.0]] m>
- __add__(particles)¶
Returns a particle subset, composed of the given particle(s) and this particle set. Attribute values are not stored by the subset. The subset provides a view on two or more sets of particles.
- Parameters
particles – (set of) particle(s) to be added to self.
>>> particles = Particles(4) >>> particles1 = particles[:2] >>> particles1.x = [1.0, 2.0] | units.m >>> particles2 = particles[2:] >>> particles2.x = [3.0, 4.0] | units.m >>> new_set = particles1 + particles2 >>> new_set <amuse.datamodel.particles.ParticlesSubset object at 0x...> >>> print len(new_set) 4 >>> print new_set.x [1.0, 2.0, 3.0, 4.0] m
- __sub__(particles)¶
Returns a subset of the set without the given particle(s) Attribute values are not stored by the subset. The subset provides a view on two or more sets of particles.
- Parameters
particles – (set of) particle(s) to be subtracted from self.
>>> particles = Particles(4) >>> particles.x = [1.0, 2.0, 3.0, 4.0] | units.m >>> junk = particles[2:] >>> new_set = particles - junk >>> new_set <amuse.datamodel.particles.ParticlesSubset object at 0x...> >>> print len(new_set) 2 >>> print new_set.x [1.0, 2.0] m >>> print particles.x [1.0, 2.0, 3.0, 4.0] m
- add_particle(particle)¶
Add one particle to the set.
- Parameters
particle – particle to add
>>> particles = Particles() >>> print len(particles) 0 >>> particle = Particle() >>> particle.x = 1.0 | units.m >>> particles.add_particle(particle) <amuse.datamodel.particles.Particle object at ...> >>> print len(particles) 1 >>> print particles.x [1.0] m
- add_particles(particles)¶
Adds particles from the supplied set to this set. Attributes and values are copied over.
Note
For performance reasons the particles are not checked for duplicates. When the same particle is part of both sets errors may occur.
- Parameters
particles – set of particles to copy values from
>>> particles1 = Particles(2) >>> particles1.x = [1.0, 2.0] | units.m >>> particles2 = Particles(2) >>> particles2.x = [3.0, 4.0] | units.m >>> particles1.add_particles(particles2) <amuse.datamodel.particles.ParticlesSubset object at 0x...> >>> print len(particles1) 4 >>> print particles1.x [1.0, 2.0, 3.0, 4.0] m
- as_set()¶
Returns a subset view on this set. The subset will contain all particles of this set.
>>> particles = Particles(3) >>> particles.x = [1.0, 2.0, 3.0] | units.m >>> subset = particles.as_set() >>> print subset.x [1.0, 2.0, 3.0] m >>> print particles.x [1.0, 2.0, 3.0] m
- copy(memento=None, keep_structure=False, filter_attributes=<function AbstractParticleSet.<lambda>>)¶
Creates a new in particle set and copies all attributes and values into this set.
The history of the set is not copied over.
Keyword arguments: memento – internal, a dictionary to keep track of already copied sets in
case of links between particles (default: None, will be created)
- keep_structure – internal, if True, in case of a sub or super set make a copy of
the original set and return a new subset (default: False)
- copy_values_of_attribute_to(attribute_name, particles)¶
Copy values of one attribute from this set to the other set. Will only copy values for the particles in both sets. See also
synchronize_to().If you need to do this a lot, setup a dedicated channel.
>>> particles1 = Particles(2) >>> particles1.x = [1.0, 2.0] | units.m >>> particles2 = particles1.copy() >>> print particles2.x [1.0, 2.0] m >>> p3 = particles1.add_particle(Particle()) >>> particles1.x = [3.0, 4.0, 5.0] | units.m >>> particles1.copy_values_of_attribute_to("x", particles2) >>> print particles2.x [3.0, 4.0] m
- difference(other)¶
Returns a new subset containing the difference between this set and the provided set.
>>> particles = Particles(3) >>> particles.mass = [10.0, 20.0, 30.0] | units.kg >>> particles.x = [1.0, 2.0, 3.0] | units.m >>> subset = particles.select(lambda x : x > 15.0 | units.kg, ["mass"]) >>> less_than_15kg = particles.difference(subset) >>> len(subset) 2 >>> len(less_than_15kg) 1
- empty_copy()¶
Creates a new in memory set and copies the particles to it. The attributes and values are not copied.The history of the set is not copied over.
>>> from amuse.datamodel import Particles >>> from amuse.units import units >>> original = Particles(2) >>> original.mass = 0 | units.m >>> print hasattr(original, "mass") True >>> print len(original) 2 >>> copy = original.empty_copy() >>> print hasattr(copy, "mass") False >>> print len(copy) 2
- ensure_presence_of(x)¶
Add one particle to the set, but only if not already in set.
- Parameters
particle – particle to add
>>> particles = Particles() >>> print len(particles) 0 >>> particle = Particle() >>> p1=particles.ensure_presence_of(particle) >>> print len(particles) 1 >>> p2=particles.ensure_presence_of(particle) >>> print len(particles) 1 >>> p1==p2 True >>> particle2 = Particle() >>> set=particle.as_set() >>> p3=set.add_particle(particle2) >>> set2=particles.ensure_presence_of(set) >>> len(particles) 2 >>> len(set2) 2
- has_duplicates()¶
Returns True when a set contains a particle with the same key more than once. Particles with the same key are interpreted as the same particles.
>>> particles = Particles() >>> p1 = particles.add_particle(Particle(1)) >>> p2 = particles.add_particle(Particle(2)) >>> particles.has_duplicates() False >>> p3 = particles.add_particle(Particle(1)) >>> particles.has_duplicates() True >>> p3 == p1 True
- remove_particle(particle)¶
Removes a particle from this set.
Result is undefined if particle is not part of the set
- Parameters
particle – particle to remove from this set
>>> particles1 = Particles(2) >>> particles1.x = [1.0, 2.0] | units.m >>> particles1.remove_particle(particles1[0]) >>> print len(particles1) 1 >>> print particles1.x [2.0] m
- remove_particles(particles)¶
Removes particles from the supplied set from this set.
- Parameters
particles – set of particles to remove from this set
>>> particles1 = Particles(2) >>> particles1.x = [1.0, 2.0] | units.m >>> particles2 = Particles() >>> particles2.add_particle(particles1[0]) <amuse.datamodel.particles.Particle object at ...> >>> particles1.remove_particles(particles2) >>> print len(particles1) 1 >>> print particles1.x [2.0] m
- reversed()¶
Returns a subset with the same particles, but with reversed sequenctial order (the first particle will become last)
>>> particles = Particles(3) >>> particles.radius = [1.0, 2.0, 3.0] | units.m >>> r = particles.reversed() >>> print r.radius [3.0, 2.0, 1.0] m
- select(selection_function, attributes)¶
Returns a subset view on this set. The subset will contain all particles for which the selection function returned True. The selection function is called with scalar quantities defined by the attributes parameter
>>> particles = Particles(3) >>> particles.mass = [10.0, 20.0, 30.0] | units.kg >>> particles.x = [1.0, 2.0, 3.0] | units.m >>> subset = particles.select(lambda x : x > 15.0 | units.kg, ["mass"]) >>> print subset.mass [20.0, 30.0] kg >>> print subset.x [2.0, 3.0] m
- select_array(selection_function, attributes=())¶
Returns a subset view on this set. The subset will contain all particles for which the selection function returned True. The selection function is called with a vector quantities containing all the values for the attributes parameter.
This function can be faster than the select function as it works on entire arrays. The selection_function is called once.
>>> particles = Particles(3) >>> particles.mass = [10.0, 20.0, 30.0] | units.kg >>> particles.x = [1.0, 2.0, 3.0] | units.m >>> subset = particles.select_array(lambda x : x > 15.0 | units.kg, ["mass"]) >>> print subset.mass [20.0, 30.0] kg >>> print subset.x [2.0, 3.0] m
>>> particles = Particles(1000) >>> particles.x = units.m.new_quantity(numpy.arange(1,1001)) >>> subset = particles.select_array(lambda x : x > (500 | units.m), ("x",) ) >>> print len(subset) 500
- sorted_by_attribute(attribute, kind='mergesort')¶
Returns a subset with the same particles, but sorted using the given attribute name
- Argument
kind, the sort method for supported kinds see the numpy.sort documentation
>>> particles = Particles(3) >>> particles.mass = [2.0, 3.0, 1.0] | units.kg >>> particles.radius = [1.0, 2.0, 3.0] | units.m >>> sorted = particles.sorted_by_attribute('mass') >>> print sorted.mass [1.0, 2.0, 3.0] kg >>> print sorted.radius [3.0, 1.0, 2.0] m
- sorted_by_attributes(*attributes)¶
Returns a subset with the same particles, but sorted using the given attribute names. The last attribute name in the call is used for the primary sort order, the second-to-last attribute name for the secondary sort order, and so on. See also numpy.lexsort
>>> particles = Particles(4) >>> particles.mass = [2.0, 3.0, 1.0, 4.0] | units.kg >>> particles.radius = [3.0, 2.0, 1.0, 2.0] | units.m >>> sorted = particles.sorted_by_attributes('mass', 'radius') >>> print sorted.radius [1.0, 2.0, 2.0, 3.0] m >>> print sorted.mass [1.0, 3.0, 4.0, 2.0] kg
- synchronize_to(other_particles)¶
Synchronize the particles of this set with the contents of the provided set.
After this call the other_particles set will have the same particles as this set.
This call will check if particles have been removed or added it will not copy values of existing particles over.
- Parameters
other_particles – particle set wich has to be updated
>>> particles = Particles(2) >>> particles.x = [1.0, 2.0] | units.m >>> copy = particles.copy() >>> new_particle = Particle() >>> new_particle.x = 3.0 | units.m >>> particles.add_particle(new_particle) <amuse.datamodel.particles.Particle object at ...> >>> print particles.x [1.0, 2.0, 3.0] m >>> print copy.x [1.0, 2.0] m >>> particles.synchronize_to(copy) >>> print copy.x [1.0, 2.0, 3.0] m
- to_string(attributes_to_show=None, split_at=20)¶
Display string of a particle set.
>>> p0 = Particle(10) >>> p1 = Particle(11) >>> particles = Particles() >>> particles.add_particle(p0) <amuse.datamodel.particles.Particle object at ...> >>> particles.add_particle(p1) <amuse.datamodel.particles.Particle object at ...> >>> particles.x = [4.0 , 3.0] | units.m >>> particles.y = [5.0 , 2.0] | units.km >>> print particles.to_string() key x y - m km ==================== =========== =========== 10 4.000e+00 5.000e+00 11 3.000e+00 2.000e+00 ==================== =========== ===========
- class amuse.datamodel.Particles(size=0, storage=None, keys=None, keys_generator=None, particles=None, is_working_copy=True, **attributes)¶
A set of particles. Attributes and values are stored in a private storage model. This storage model can store the values in the python memory space, in the memory space of the code or in a HDF5 file. By default the storage model is in memory.
- class amuse.datamodel.ParticlesSubset(particles, keys)¶
A subset of particles. Attribute values are not stored by the subset. The subset provides a limited view to the particles.
Particle subset objects are not supposed to be created directly. Instead use the
to_setorselectmethods.- add_particles_to_store(keys, attributes=[], values=[])¶
Adds particles from to the subset, also adds the particles to the superset
- as_set()¶
Returns a subset view on this set. The subset will contain all particles of this set.
>>> particles = Particles(3) >>> particles.x = [1.0, 2.0, 3.0] | units.m >>> subset = particles.as_set() >>> print subset.x [1.0, 2.0, 3.0] m >>> print particles.x [1.0, 2.0, 3.0] m
- copy(memento=None, keep_structure=False, filter_attributes=<function ParticlesSubset.<lambda>>)¶
Creates a new in particle set and copies all attributes and values into this set.
The history of the set is not copied over.
Keyword arguments: memento – internal, a dictionary to keep track of already copied sets in
case of links between particles (default: None, will be created)
- keep_structure – internal, if True, in case of a sub or super set make a copy of
the original set and return a new subset (default: False)
- difference(other)¶
Returns a new subset containing the difference between this set and the provided set.
>>> particles = Particles(3) >>> particles.mass = [10.0, 20.0, 30.0] | units.kg >>> particles.x = [1.0, 2.0, 3.0] | units.m >>> subset = particles.select(lambda x : x > 15.0 | units.kg, ["mass"]) >>> less_than_15kg = particles.difference(subset) >>> len(subset) 2 >>> len(less_than_15kg) 1
- remove_particles_from_store(indices)¶
Removes particles from the subset, and removes particles from the super set
- union(other)¶
Returns a new subset containing the union between this set and the provided set.
>>> particles = Particles(3) >>> particles.mass = [10.0, 20.0, 30.0] | units.kg >>> subset1 = particles.select(lambda x : x > 25.0 | units.kg, ["mass"]) >>> subset2 = particles.select(lambda x : x < 15.0 | units.kg, ["mass"]) >>> union = subset1.union(subset2) >>> len(union) 2 >>> sorted(union.mass.value_in(units.kg)) [10.0, 30.0]
- class amuse.datamodel.ParticlesWithUnitsConverted(particles, converter)¶
A view on a particle sets. Used when to convert values between incompatible sets of units. For example to convert from si units to nbody units.
The converter must have implement the ConverterInterface.
>>> from amuse.units import nbody_system >>> particles_nbody = Particles(2) >>> particles_nbody.x = [10.0 , 20.0] | nbody_system.length >>> convert_nbody = nbody_system.nbody_to_si(10 | units.kg , 5 | units.m ) >>> particles_si = ParticlesWithUnitsConverted( ... particles_nbody, ... convert_nbody.as_converter_from_si_to_nbody()) ... >>> print particles_nbody.x [10.0, 20.0] length >>> print particles_si.x [50.0, 100.0] m >>> particles_si.x = [200.0, 400.0] | units.m >>> print particles_nbody.x [40.0, 80.0] length
- class ConverterInterface¶
Interface definition for the converter.
- source
The source quantity is in the units of the user of a ParticlesWithUnitsConverted object
- target
The target quantity must be in the units of the internal particles set.
- from_source_to_target()¶
Converts the quantity from the source units to the target units.
- Parameters
quantity – quantity to convert
- from_target_to_source()¶
Converts the quantity from the target units to the source units.
- Parameters
quantity – quantity to convert
- as_set()¶
Returns a subset view on this set. The subset will contain all particles of this set.
>>> particles = Particles(3) >>> particles.x = [1.0, 2.0, 3.0] | units.m >>> subset = particles.as_set() >>> print subset.x [1.0, 2.0, 3.0] m >>> print particles.x [1.0, 2.0, 3.0] m
object¶
- class amuse.datamodel.Particle(key=None, particles_set=None, set_index=None, set_version=-1, **keyword_arguments)¶
A physical object or a physical region simulated as a physical object (cloud particle).
All attributes defined on a particle are specific for that particle (for example mass or position). A particle contains a set of attributes, some attributes are generic and applicable for multiple modules. Other attributes are specific and are only applicable for a single module.
- __add__(particles)¶
Returns a particle subset, composed of the given particle(s) and this particle. Attribute values are not stored by the subset. The subset provides a view on the particles.
- Parameters
particles – particle(s) to be added to self.
>>> particles = Particles(2) >>> particle1 = particles[0] >>> particle1.x = 1.0 | units.m >>> particle2 = particles[1] >>> particle2.x = 2.0 | units.m >>> new_set = particle1 + particle2 >>> new_set <amuse.datamodel.particles.ParticlesSubset object at 0x...> >>> print len(new_set) 2 >>> print new_set.x [1.0, 2.0] m
- __sub__(particles)¶
Raises an exception: cannot subtract particle(s) from a particle.
- as_set()¶
Returns a subset view on the set containing this particle. The subset view includes this particle and no other particles.
>>> particles = Particles(2) >>> particles.x = [1.0, 2.0] | units.m >>> particle2 = particles[1] >>> print particle2.x 2.0 m >>> particles_with_one_particle = particle2.as_set() >>> len(particles_with_one_particle) 1 >>> print particles_with_one_particle.x [2.0] m
Methods to retreive physical properties of the particles set¶
- amuse.datamodel.particle_attributes.center_of_mass(particles)
Returns the center of mass of the particles set. The center of mass is defined as the average of the positions of the particles, weighted by their masses.
>>> from amuse.datamodel import Particles >>> particles = Particles(2) >>> particles.x = [-1.0, 1.0] | units.m >>> particles.y = [0.0, 0.0] | units.m >>> particles.z = [0.0, 0.0] | units.m >>> particles.mass = [1.0, 1.0] | units.kg >>> particles.center_of_mass() quantity<[0.0, 0.0, 0.0] m>
- amuse.datamodel.particle_attributes.center_of_mass_velocity(particles)
Returns the center of mass velocity of the particles set. The center of mass velocity is defined as the average of the velocities of the particles, weighted by their masses.
>>> from amuse.datamodel import Particles >>> particles = Particles(2) >>> particles.vx = [-1.0, 1.0] | units.ms >>> particles.vy = [0.0, 0.0] | units.ms >>> particles.vz = [0.0, 0.0] | units.ms >>> particles.mass = [1.0, 1.0] | units.kg >>> particles.center_of_mass_velocity() quantity<[0.0, 0.0, 0.0] m * s**-1>
- amuse.datamodel.particle_attributes.kinetic_energy(particles)
Returns the total kinetic energy of the particles in the particles set.
>>> from amuse.datamodel import Particles >>> particles = Particles(2) >>> particles.vx = [-1.0, 1.0] | units.ms >>> particles.vy = [0.0, 0.0] | units.ms >>> particles.vz = [0.0, 0.0] | units.ms >>> particles.mass = [1.0, 1.0] | units.kg >>> particles.kinetic_energy() quantity<1.0 m**2 * kg * s**-2>
- amuse.datamodel.particle_attributes.potential_energy(particles, smoothing_length_squared=quantity<zero>, G=quantity<6.67428e-11 m**3 * kg**-1 * s**-2>)
Returns the total potential energy of the particles in the particles set.
- Parameters
smooting_length_squared – gravitational softening, added to every distance**2.
G – gravitational constant, need to be changed for particles in different units systems
>>> from amuse.datamodel import Particles >>> particles = Particles(2) >>> particles.x = [0.0, 1.0] | units.m >>> particles.y = [0.0, 0.0] | units.m >>> particles.z = [0.0, 0.0] | units.m >>> particles.mass = [1.0, 1.0] | units.kg >>> particles.potential_energy() quantity<-6.67428e-11 m**2 * kg * s**-2>
- amuse.datamodel.particle_attributes.particle_specific_kinetic_energy(set, particle)
Returns the specific kinetic energy of the particle.
>>> from amuse.datamodel import Particles >>> particles = Particles(2) >>> particles.vx = [0.0, 1.0] | units.ms >>> particles.vy = [0.0, 0.0] | units.ms >>> particles.vz = [0.0, 0.0] | units.ms >>> particles.mass = [1.0, 1.0] | units.kg >>> particles[1].specific_kinetic_energy() quantity<0.5 m**2 * s**-2>
- amuse.datamodel.particle_attributes.particle_potential(set, particle, smoothing_length_squared=quantity<zero>, G=quantity<6.67428e-11 m**3 * kg**-1 * s**-2>)
Returns the potential at the position of the particle.
- Parameters
smooting_length_squared – gravitational softening, added to every distance**2.
G – gravitational constant, need to be changed for particles in different units systems
>>> from amuse.datamodel import Particles >>> particles = Particles(2) >>> particles.x = [0.0, 1.0] | units.m >>> particles.y = [0.0, 0.0] | units.m >>> particles.z = [0.0, 0.0] | units.m >>> particles.mass = [1.0, 1.0] | units.kg >>> particles[1].potential() quantity<-6.67428e-11 m**2 * s**-2>
Implementation¶
![digraph multiples {
fontsize=8.0;
node [fontsize=8.0,shape=box, style=filled, fillcolor=lightyellow];
"AttributeStorage" -> "AbstractSet";
"AbstractSet" -> "AbstractParticlesSet";
"AbstractSet" -> "AbstractGrid";
"AbstractParticlesSet" -> "Particles";
"AbstractParticlesSet" -> "ParticlesSubSet";
"AbstractParticlesSet" -> "ParticlesSuperSet";
"AbstractGrid" -> "Grid";
"AbstractGrid" -> "SubGrid";
"AttributeStorage" -> "InMemoryStorage" [label = "implements"];
"AttributeStorage" -> "InCodeStorage";
}](../_images/graphviz-7cc5ca3f72add54679248f70b0787cd0f680b296.png)
![digraph multiples {
fontsize=8.0;
node [fontsize=8.0,shape=box, style=filled, fillcolor=lightyellow];
edge [color="dodgerblue2", fontcolor="dodgerblue2"];
"Particles" -> "AttributeStorage" [headlabel=" 1", taillabel="1",label = "store"];
"Grid" -> "AttributeStorage" [headlabel=" 1", taillabel="1", label = "store"];
"SubGrid" -> "Grid" [headlabel=" 1", taillabel="1", label="view on", color="dodgerblue2"];
"ParticlesSubSet" -> "Particles" [headlabel=" 1", taillabel="1", label="view on", color="dodgerblue2"];
"ParticlesSuperSet" -> "Particles" [headlabel=" *", taillabel="1", label="view on", color="dodgerblue2"];
}](../_images/graphviz-f8a9687a4a009a6bd4f961385fb780b9e46a8a6a.png)
![digraph multiples {
fontsize=8.0;
node [fontsize=8.0,shape=box, style=filled, fillcolor=lightyellow];
"AbstractSet" -> "AttributeStorage" [headlabel=" 1", taillabel="1", label = "stored_attributes", color="dodgerblue2" , fontcolor="dodgerblue2"];
"AbstractSet" -> "DerivedAttribute" [headlabel=" *", taillabel="1", label = "derived_attributes", color="dodgerblue2", fontcolor="dodgerblue2"];
"DerivedAttribute" -> "CalculatedAttribue";
"DerivedAttribute" -> "VectorAttribute";
"DerivedAttribute" -> "FunctionAttribute";
"DerivedAttribute" -> "DomainAttribute";
}](../_images/graphviz-9d90cd40ae28ccb57a151a769454bb00a9eaf00d.png)
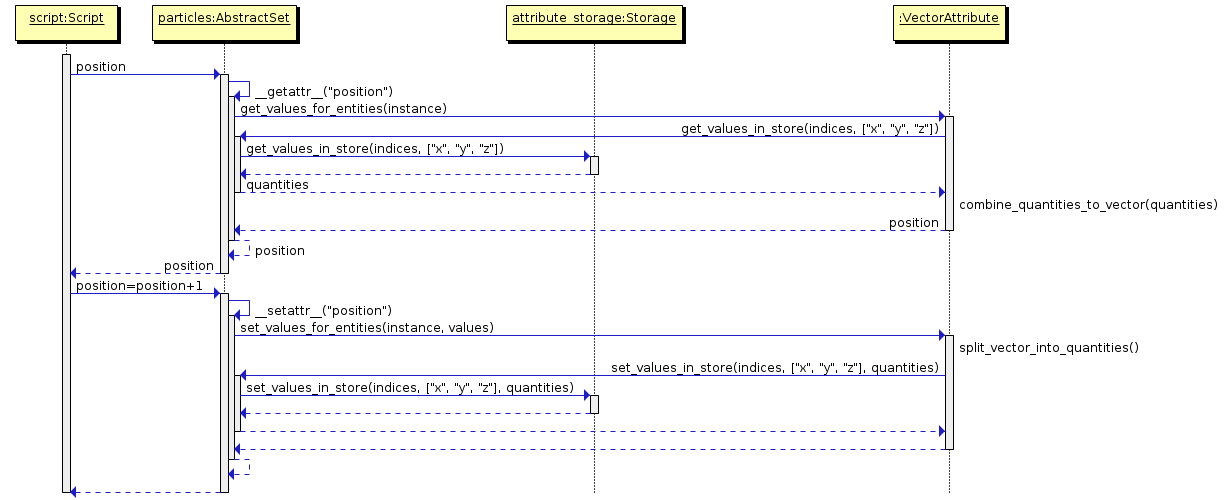
- class amuse.datamodel.AttributeStorage¶
Abstract base class of particle storage objects. Implemented by the storage classes and the particle sets
- add_particles_to_store(keys, attributes=[], values=[])¶
Adds particles with the given keys to the set. If attribute names and values are specified these are also set for the new particles.
- get_all_indices_in_store()¶
Gets the list of all valid indices in store.
- get_all_keys_in_store()¶
Gets the list of all keys stored
- get_attribute_names_defined_in_store()¶
Gets the attribute names
- get_indices_of_keys(keys)¶
Returns the indices where the particles are stored with the given keys.
- get_value_in_store(index, attribute)¶
Returns the value of an attribute for a particle at the given index.
- get_values_in_store(indices, attributes)¶
Gets the values for the attributes of the particles at the given indices.
- has_key_in_store(key)¶
Returns true if the given key can be found in the store
- remove_particles_from_store(keys)¶
Removes particles with the given keys from the set.
- set_values_in_store(indices, attributes, list_of_values_to_set)¶
Sets the attributes of the particles at the given indices to the given values.